Meet GeoFlow V3, BioGeometry’s next-generation all-atom foundation model for protein design.
For the first time, GeoFlow V3 unifies generation, evaluation and in silico evolution of antibody binders, significantly improving their bind rates to 15.5%, 50x higher than SOTA methods in 2024. Validated on 8 de novo design campaigns, GeoFlow V3 routinely discovers nanomolar hits within 50 tested designs and 3 weeks of time.
Its high hit rates and precision is pushing the boundary of AI-driven de novo antibody design.
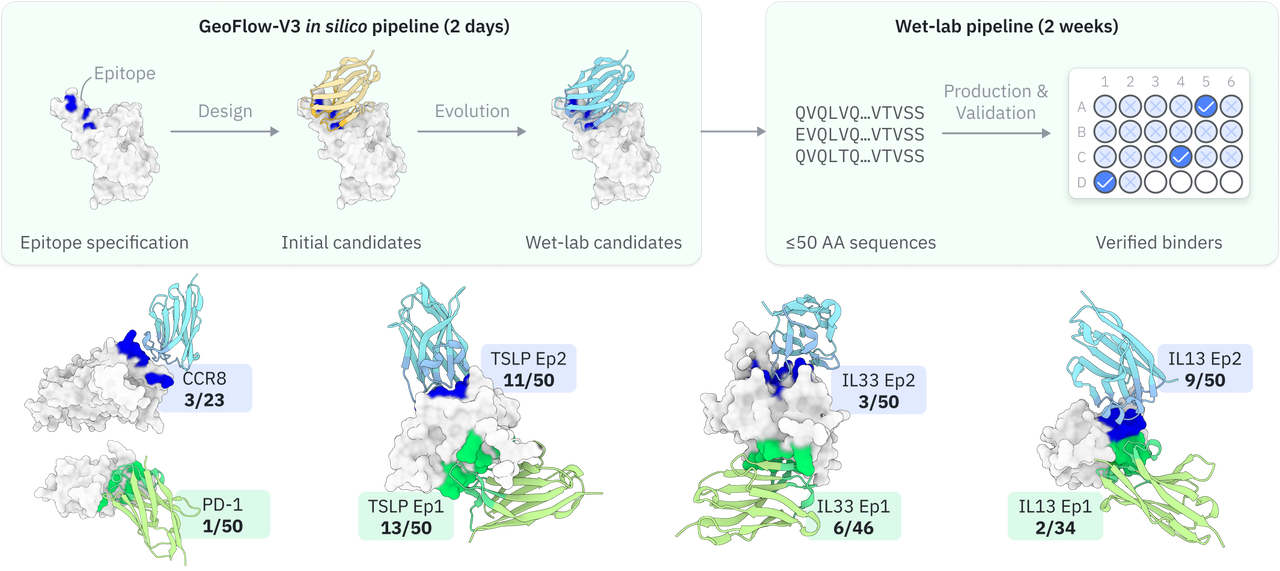
An atomic diffusion model for “epitope-in-antibody-out” design
Building on GeoFlow V2, the first all-atom model to unify structure prediction and de novo design, GeoFlow V3 takes the next step: it replaces the common “generate-filter” workflow with a “design–evolve” loop, allowing the model to iteratively refine its own designs. The improved architecture, training and inference scheme lead to improved accuracy, reliability, and overall hit rates.
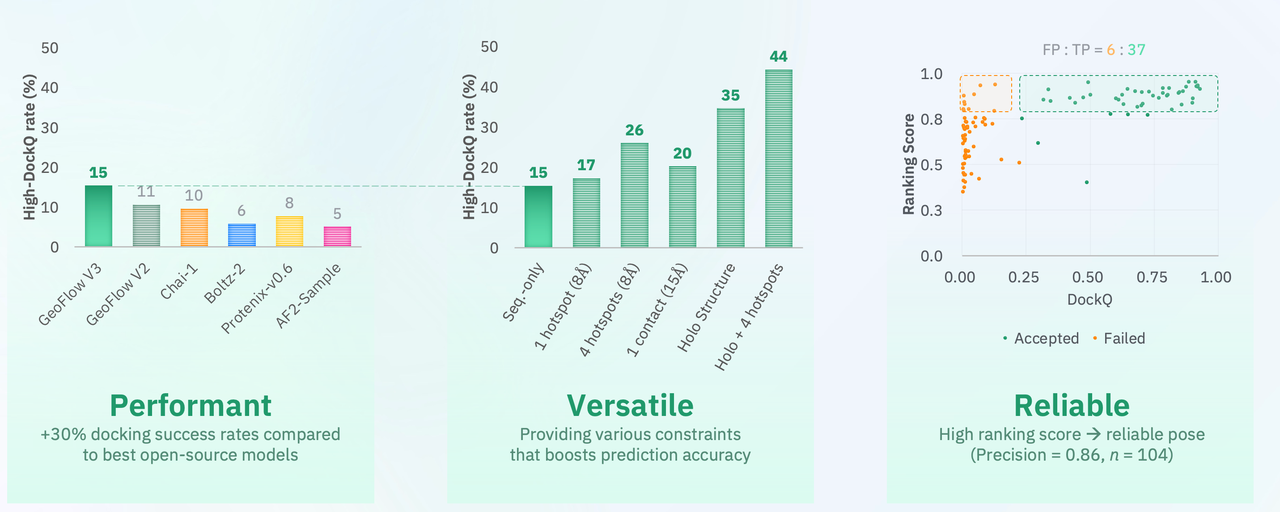
Predicting high-quality atomic structures
GeoFlow V3 improves high-DockQ rate by 45% compared with its predecessor, GeoFlow V2. The model can incorporate epitope conditioning and structural cues when available, which further boosts prediction accuracy.
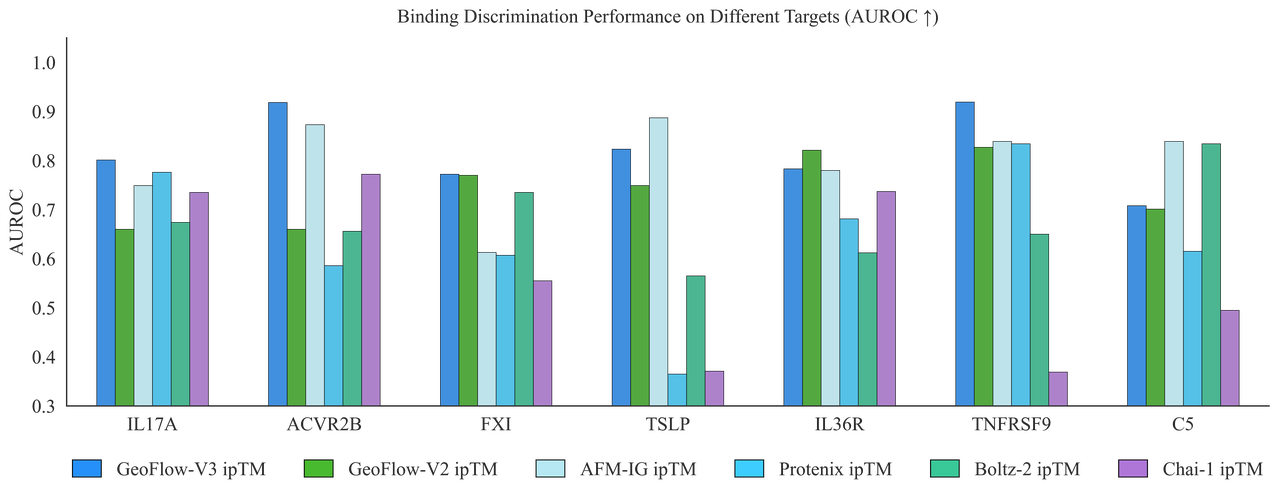
Distinguishing binders from non-binders
GeoFlow V3 also shows marked improvement in self-evaluation capabilities. Among benchmarked methods, it is the most robust in distinguishing binders from non-binders. This gives GeoFlow V3 the ability to identify not only promising candidates but also “hotspots” within them which, after redesign, could further improve affinity.
Simulating evolution in silico
Inspired by natural affinity maturation, GeoFlow V3 introduces in silico evolution—a process where candidate antibodies undergo multi-round refinement guided by internal confidence scores.
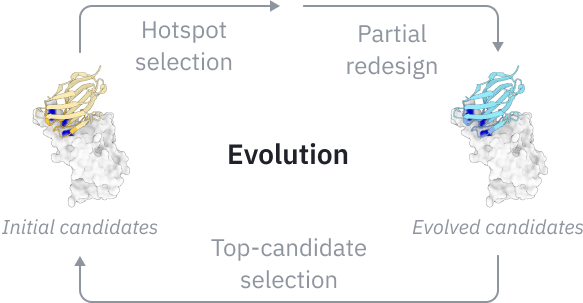
This iterative process significantly improves ipTM scores, a key indicator of binding potential, showing that scaling inference-time compute can yield stronger molecular interactions and save wet-lab costs.
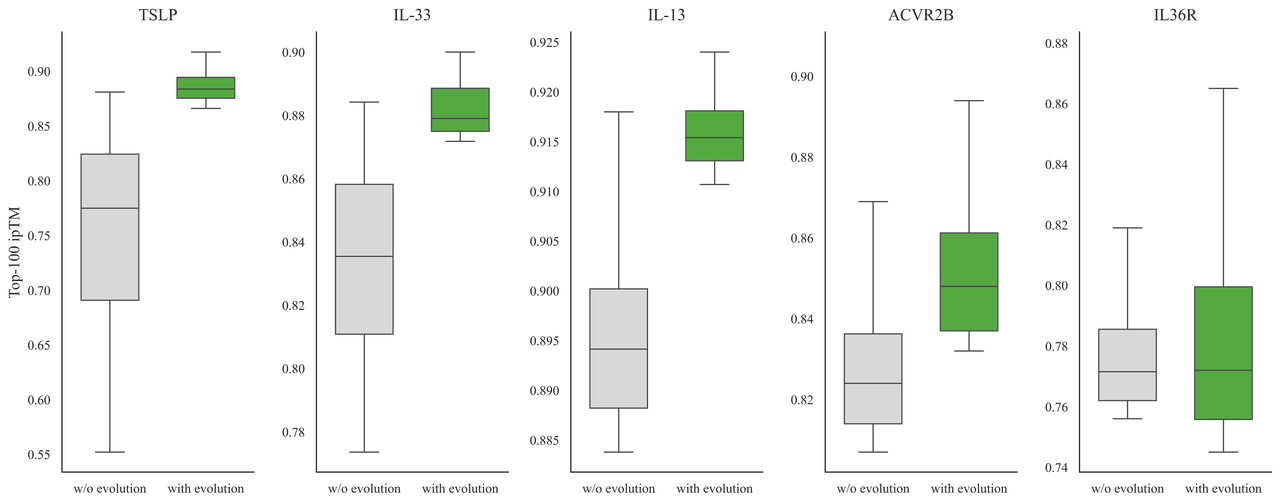
More hits in fewer tests
Traditional antibody discovery relies on massive molecular libraries and months of screening – either in vivo through animal immunization, or in vitro through library screening. Even the best AI tools in 2024 must be coupled with library construction and screening to reliably get hits, as their hit rate were often below 1%.
With GeoFlow V3, we no longer need months for design validation.
In tests across eight epitopes on five therapeutically relevant targets—TSLP, IL-13, IL-33, CCR8, and PD-1, GeoFlow V3 identified binders within 50 tested candidates and 3 weeks time. The average wet-lab hit rate reaches 15.5%, dozens of times higher than previous generation models.
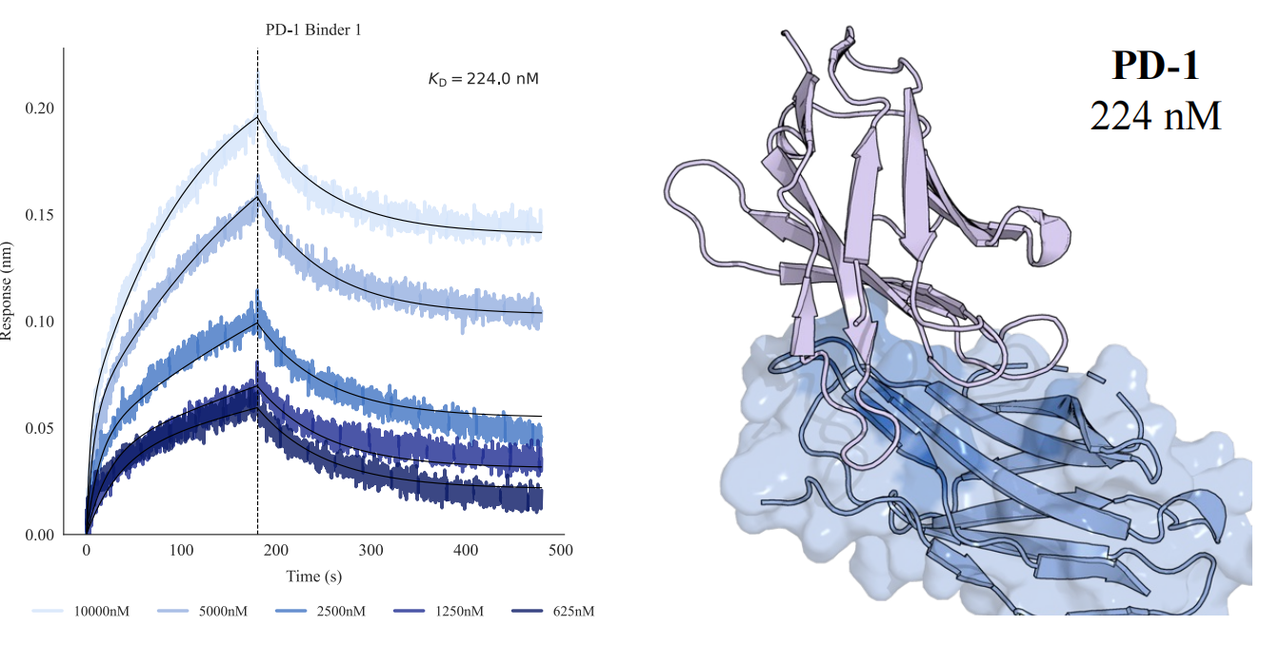
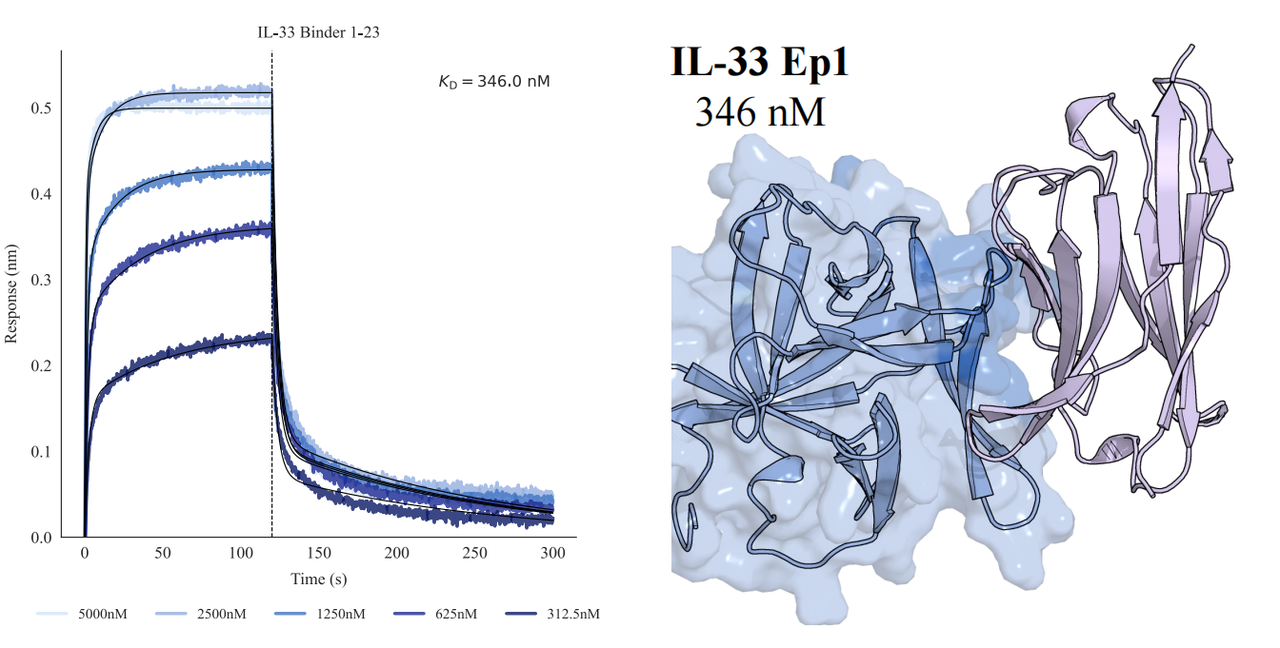
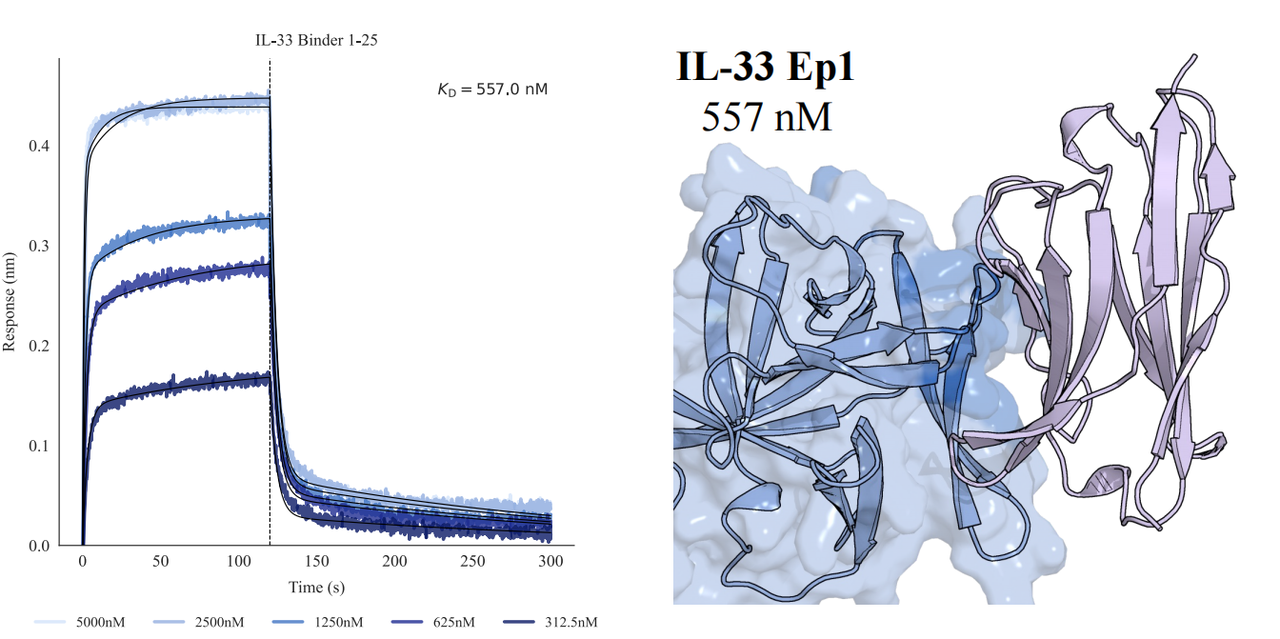
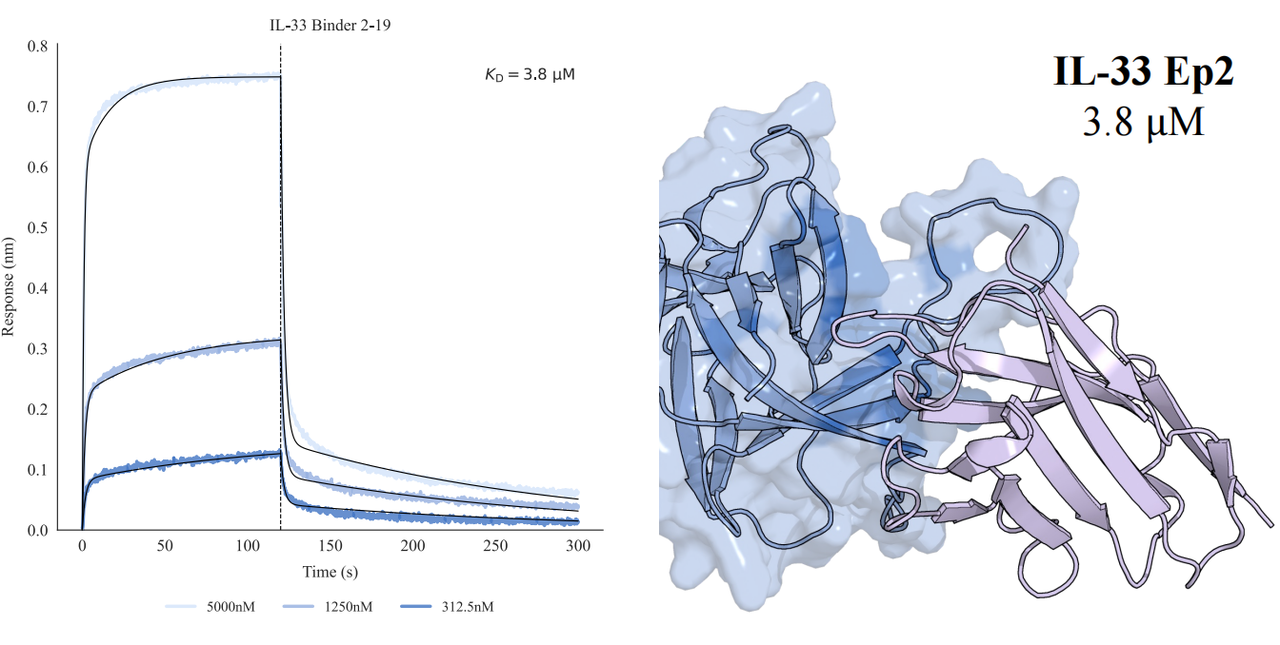
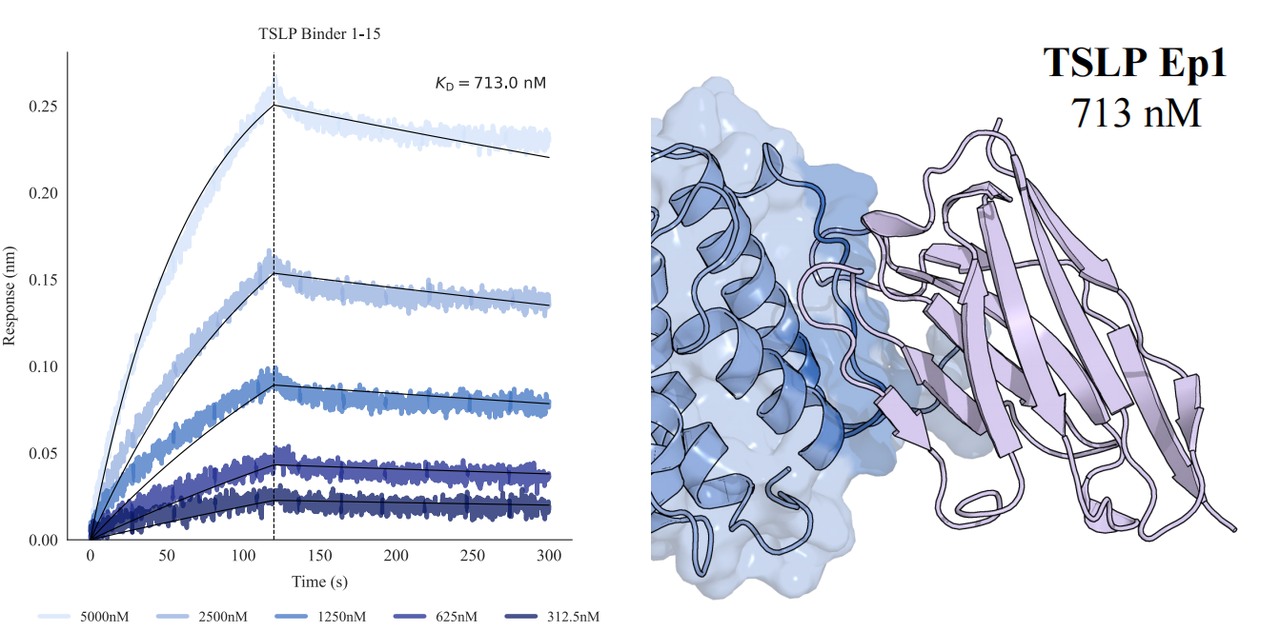
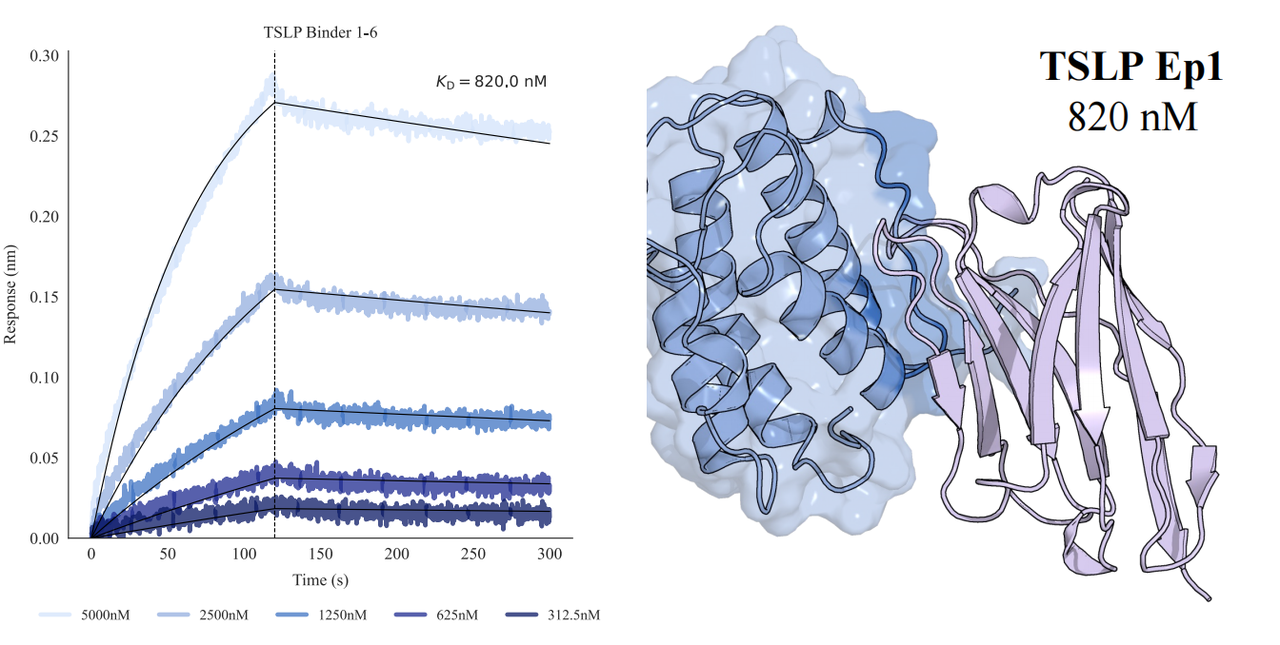
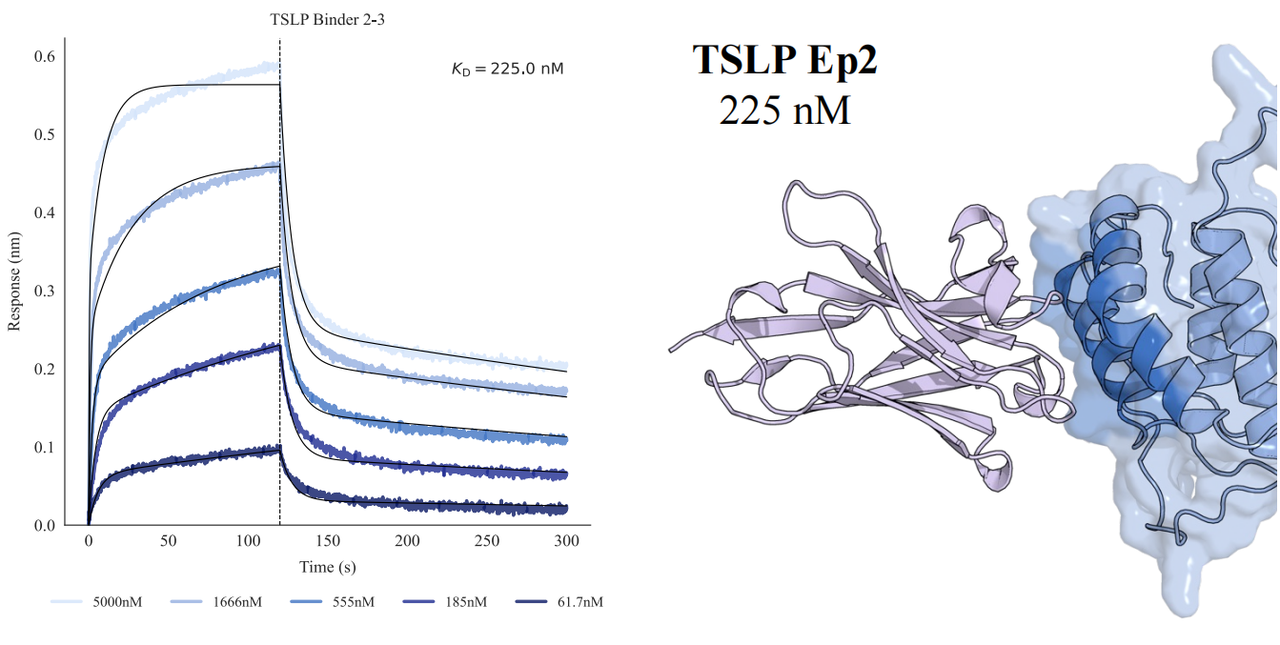
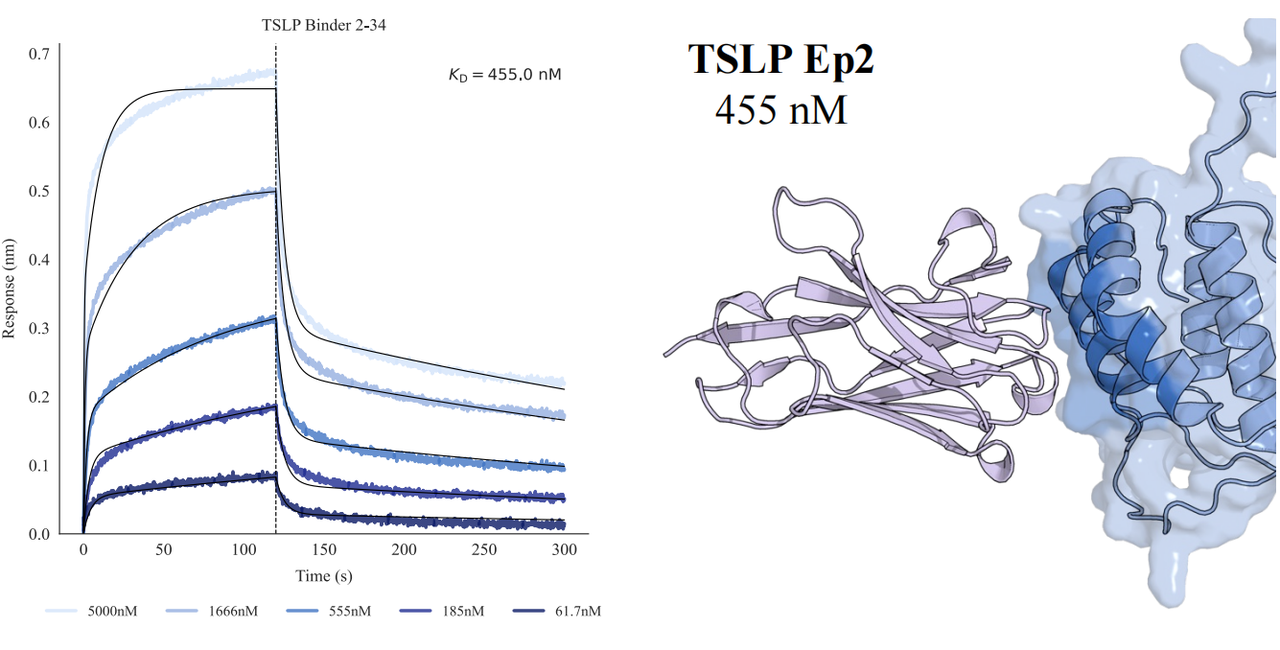
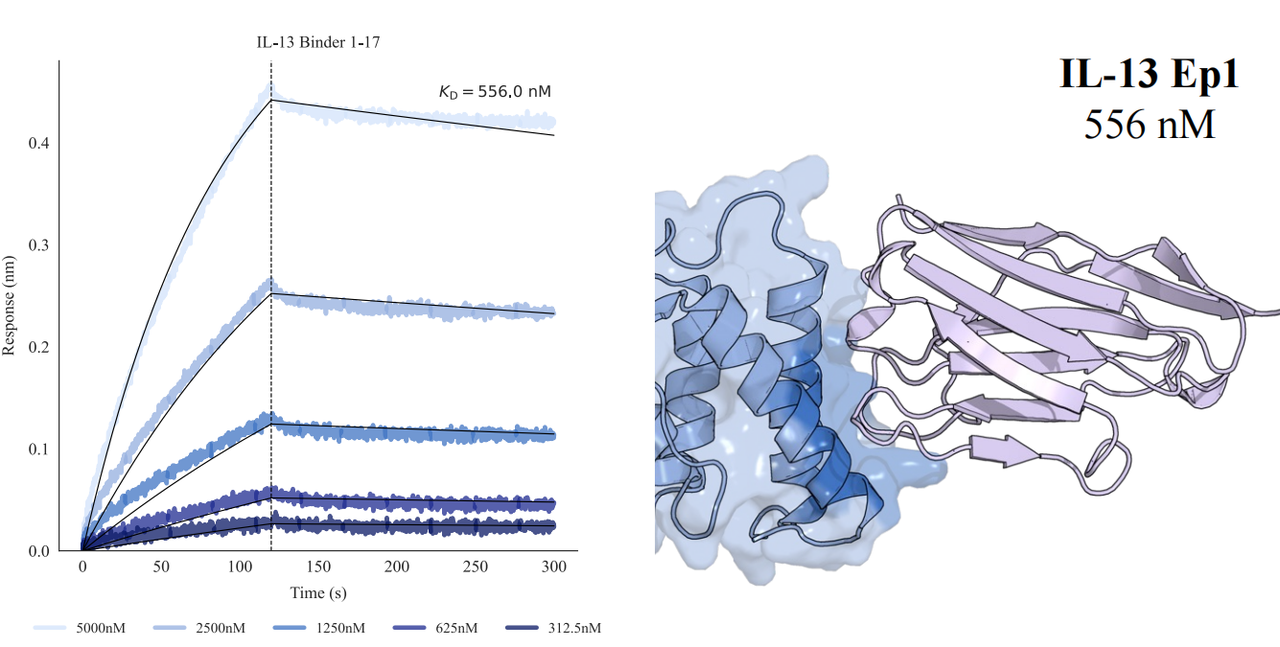
Put simply, dozens of designs—not tens of thousands—are now enough to find viable, diverse hits. With AI relieving the burden of hit discovery, researchers can focus more on discovering functional epitopes and developing promising molecules.
A unified engine for biomedicine and biomanufacturing
GeoFlow V3 isn’t just for antibodies — it is pushing the frontier of protein design and optimization.

In biomedicine, the model shortens the path from concept to candidate, cutting hit discovery times from months to less than three weeks. It enables targeting of epitopes previously considered undruggable, bringing precision design into areas long driven by trial and error.
In biomanufacturing, atomic-level enzyme design enables sustainable, cost-effective bio-production for food, supplements, cosmetics, fragrances, etc.
GeoFlow V3 is dramatically accelerating the translation of scientific discovery into industrial applications of proteins. It converts what used to be a stochastic, empirical search into a precise, programmable engineering solution.
